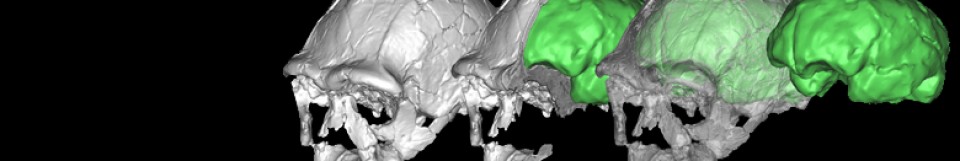
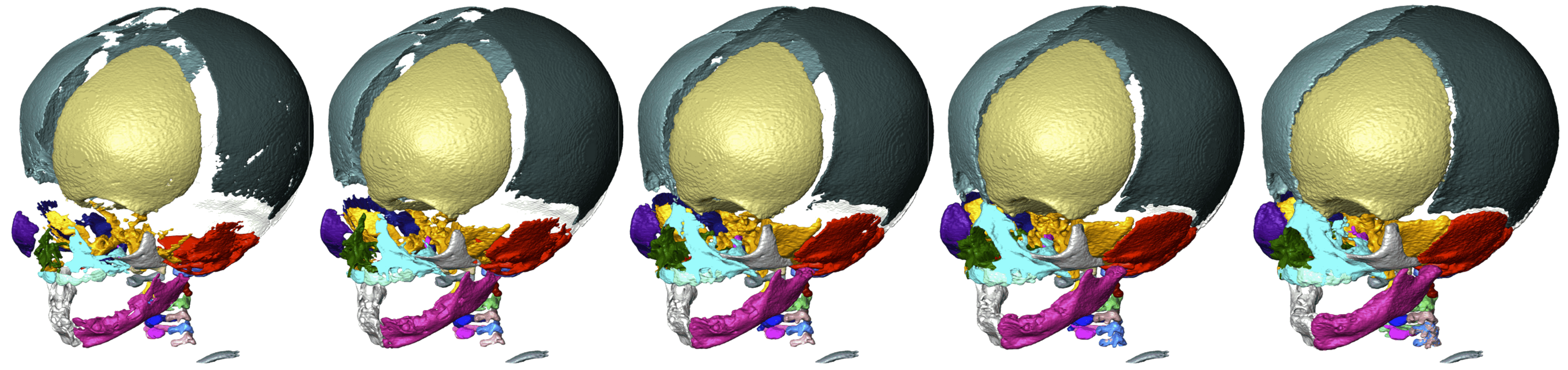Computed tomography (CT) scanning is a standard way of visualizing hard tissues in living organisms. However, tridimensional reconstruction of CT images requires segmenting the structure at hand, which is time-consuming at best (i.e., manual segmentation) and imprecise at worst (i.e., automatic segmentation), especially in multipart segmentation. To circumvent this issue, Didziokas et al. (2024) have developed an open-access, user-friendly, automated segmentation tool for hard tissues, focusing especially on skull bones: boundary-preserving threshold iteration (BounTI). As its name suggests, BounTI’s operators select the structure of interest based on voxel intensity, which is the only input parameter it needs from the user. This procedure yields good results for bone segmentation, given that osseous tissue usually presents a distinct voxel intensity when compared with its surroundings. An appropriate initial threshold of voxel intensity is one that does not cause separate elements to be joint in the seed (that is, the first recognition of tissue by the algorithm), and which does not cause erroneous separations of single elements (e.g., the parietal bone) in the final stage.
BounTI was tested on skull CT images of various species, including amphibians, reptiles, and mammals. The quality of the assessment’s results demonstrates BounTI’s versatility and effectiveness. However, its performance is bound by the quality of the image; lower resolutions yield worse results. To mend any errors that might arise, BounTI does include options for manual intervention. Lastly, the authors emphasize the tool’s accessibility, human and machine-wise. BounTI can be implemented in a plethora of ways, holding great potential to improve efficiency and accuracy in anatomical studies and clinical applications involving hard tissue segmentation.
Tim Schuurman